People • Publications • Research • Contact • Links
We study the relationship between the molecular, cellular, and developmental mechanisms that generate phenotypes and the population- and community-level factors that shape variation and evolution of those phenotypes. This is evo-devo. Our primary tool is experimental genetics, and our primary experimental organisms are nematodes and polychaetes. A few themes are outlined below.
What are the alleles underlying phenotypic variation in wild populations? We use experimental genetics in Caenorhabditis nematodes to investigate the genetic basis for phenotypic variation. One current project uses experimental evolution to discover the characteristics of recessive deleterious variants that segregate in wild populations of an obligately outcrossing species. We aim to develop models that let us predict dominance coefficients from sequence. A second active project uses comparative quantitative genetics in multiple species to address the evolution of genetic architecture of resistance to environmental toxins.
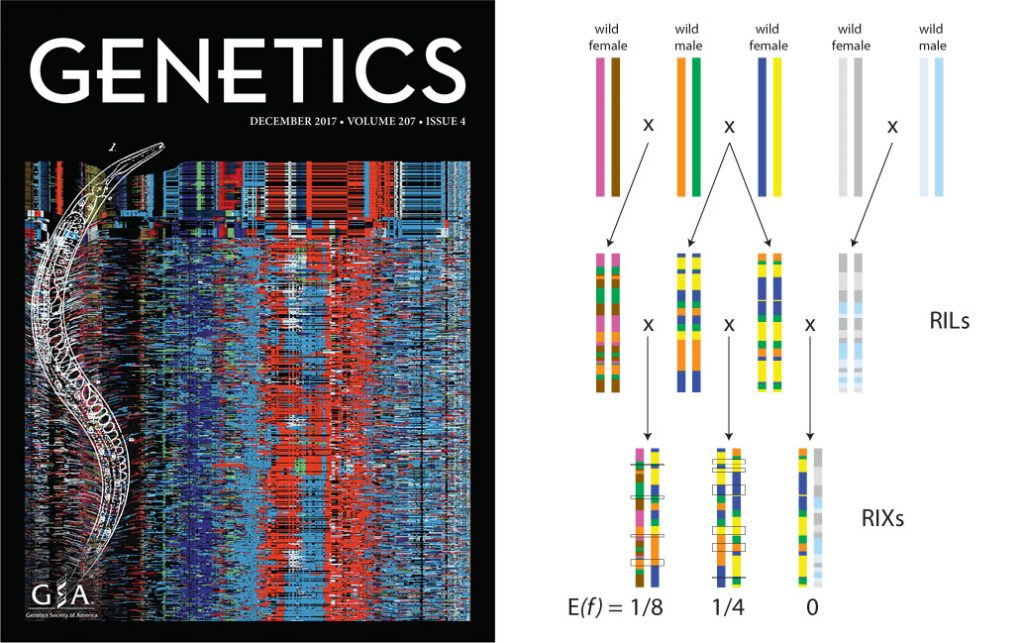
Left: the C. elegans Multiparent Experimental Evolution Panel. Right: our C. becei RILs allow us to manipulate autozygosity while controlling allele frequencies.
How and why does early development evolve? We study a marine annelid, Streblospio benedicti, that exhibits dramatic variation in embryonic and larval phenotypes. This species gives us a genetic handle on questions at the intersection of life-history theory, population genomics, and developmental biology. We are currently working to connect the unique transmission-genetic issues of early development (e.g., maternal genetic effects) with the empirical patterns of genetic and phenotypic variation in S. benedicti.
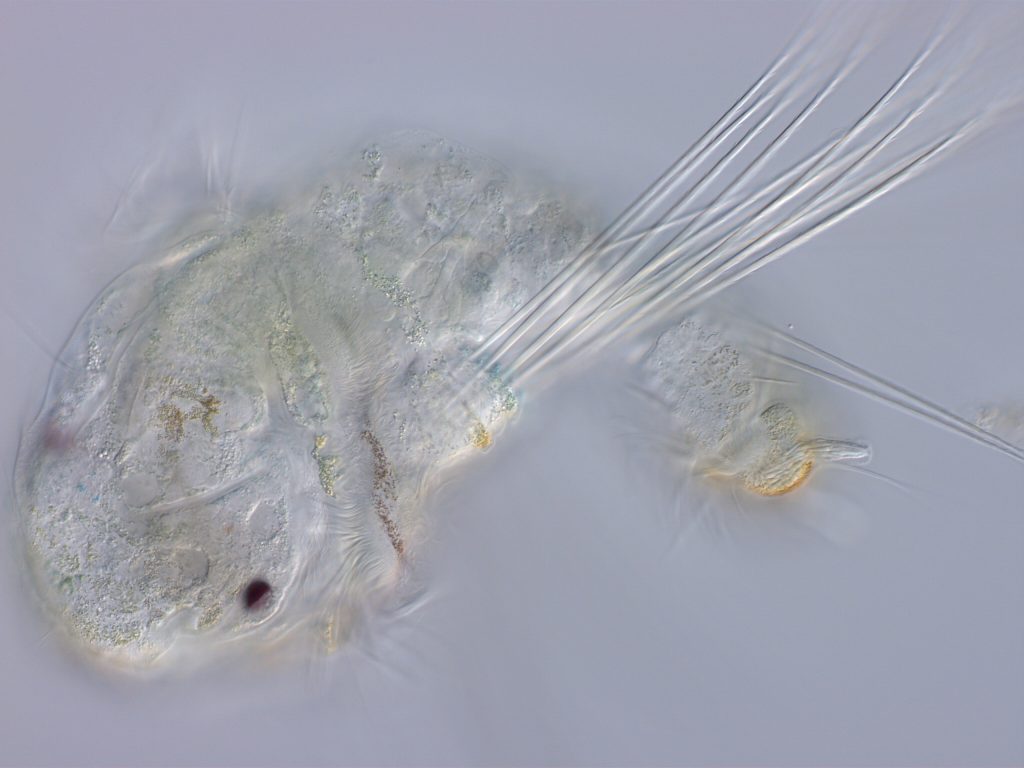
A planktotrophic larva of S. benedicti. Lecithotrophic larvae are eight times larger and lack chaetae and cirri.
How does the habitat of Caenorhabditis nematodes shape their biology? C. elegans and its relatives are real animals that live in nature. We are working to understand how the animals are adapted to their native conditions of ephemeral habitat patches, using theory, lab experiments, and field data to address classical evolutionary questions. In the process we are discovering new species and contributing to basic biogeographic and ecological knowledge about these worms.

Left: Tracking Caenorhabditis on Barro Colorado Island, Panamá. Right: the distinctive male tail of C. sp. 52, a new species from the rainforests of Queensland’s Main Range.
How does linkage shape phenotypic evolution? The fate of an allele in a population depends on how independently it is transmitted from generation to generation. Rates and patterns of sex and recombination govern this allelic independence. We study how linkage influences the pattern of trait variation and covariation among nematode species that vary in their rates of outcrossing and among genomic regions that vary in their rates of recombination. In parallel we ask how outcrossing and recombination evolve.
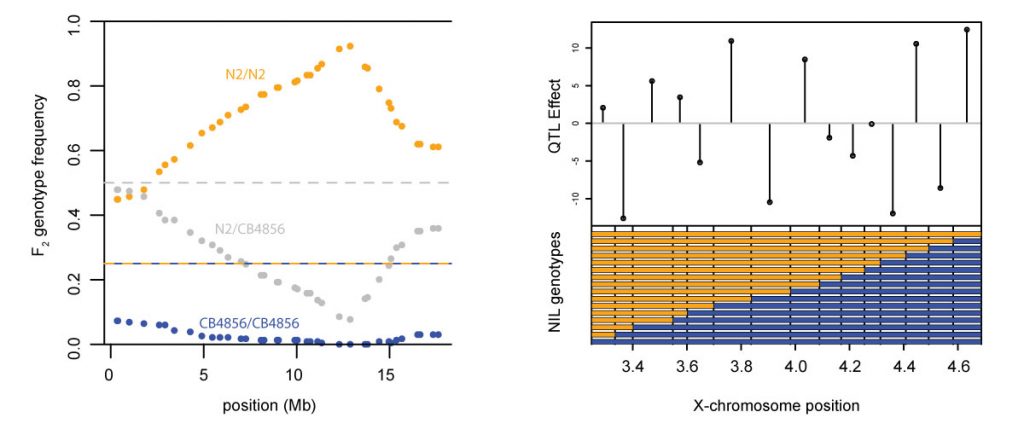
Left: strong selection on a single locus distorts allele frequencies across an entire chromosome. Right: Tight linkage between loci of opposite effects suppresses phenotypic variation.